CytoSpec - an APPLICATION FOR HYPERSPECTRAL IMAGING |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||







|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
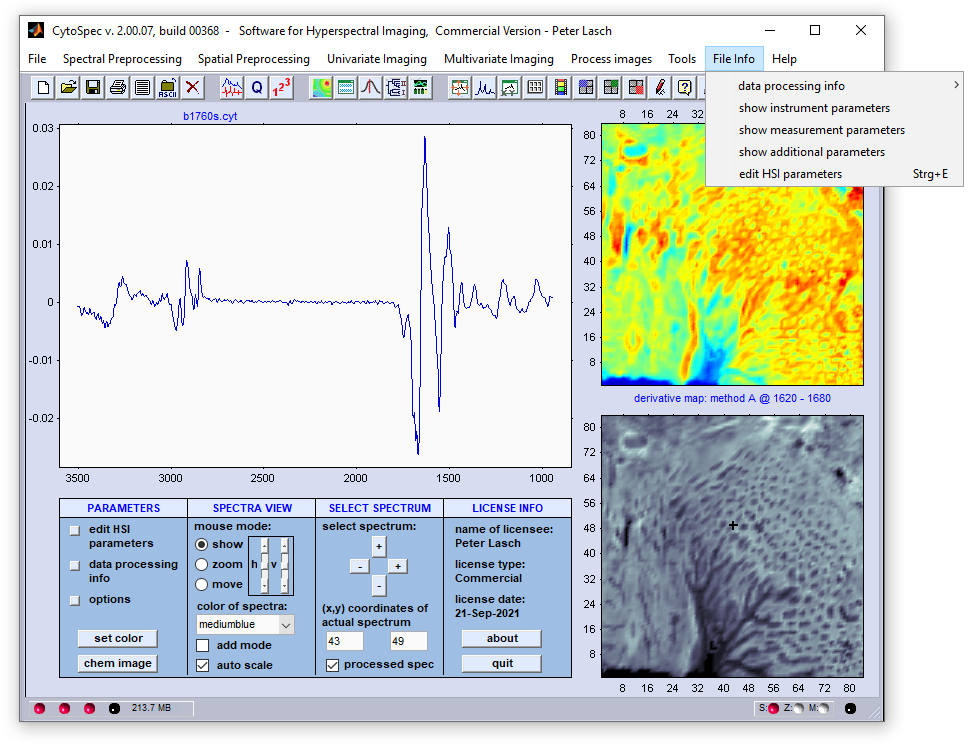
Menu Bar 'File Info' |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Show Data Block History |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
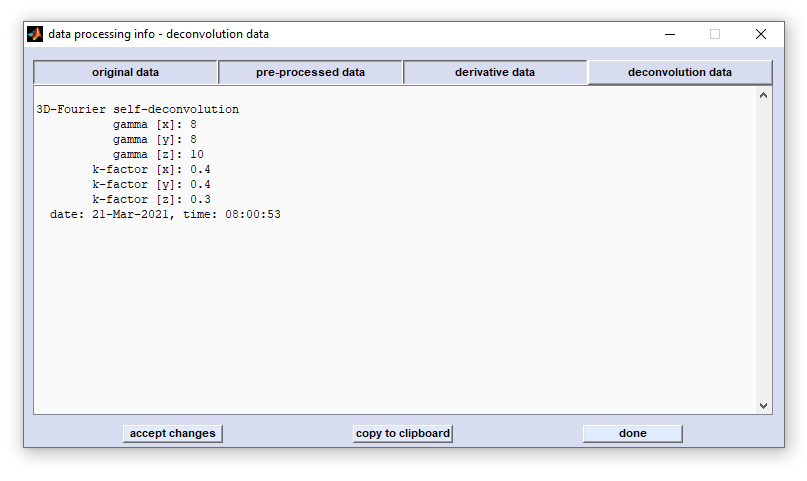
Edit Parameters and HSI Metadata |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
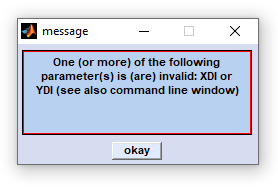
define the spectral vector and thus represent internal program-wide variables that can have variety of effects when changes are made. The same applies, in principle, to XDI and YDI which denote the number of pixel spectra in x-, and y-direction of the given HSI (see below) for details. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Instrument parameters: | Measurement parameters: | Additional data acquisition parameters: | |||
| INS | - instrument type | LWN | - starting wavenumber | TOS | - type of spectra |
| MAN | - instrument manufacturer | UWN | - ending wavenumber | NOS | - number of scans |
| SRC | - source setting | WVS | - wavenumber step | XDI | - number of pixel in x-direction |
| BSP | - beamsplitter setting | NOD | - number of spectral data points | YDI | - number of pixel in y-direction |
| FIL | - type of optical filter | DAT | - date of measurement | NSP | - number of spectra |
| DET | - type of detector | TIM | - time of measurement | SZX | - area size in x dimension (µm) |
| VEL | - scanner velocity | PHC | - phase correction mode | SZY | - area size in y dimension (µm) |
| LAS | - laser wavenumber | PHR | - phase resolution | SAM | - sample name |
| AQM | - acquisition mode | PIP | - phase interferogram points | SAF | - sample form |
| SNG | - signal gain | ZFF | - zero filling factor | OPN | - operator name |
| RES | - nominal resolution | AS1 | - additional sample information 1 | ||
| APO | - apodization function | AS2 | - additional sample information 2 | ||
| APT | - aperture size (µm) | AS3 | - additional sample information 3 | ||
| STY | - step in y-dimension (µm) | ||||
| STX | - step in x-dimension (µm) | ||||
|
Parameters highlighted by red color should be carefully modified since these parameters are used by
the CytoSpec software. Modification of these parameters may cause malfunctioning.
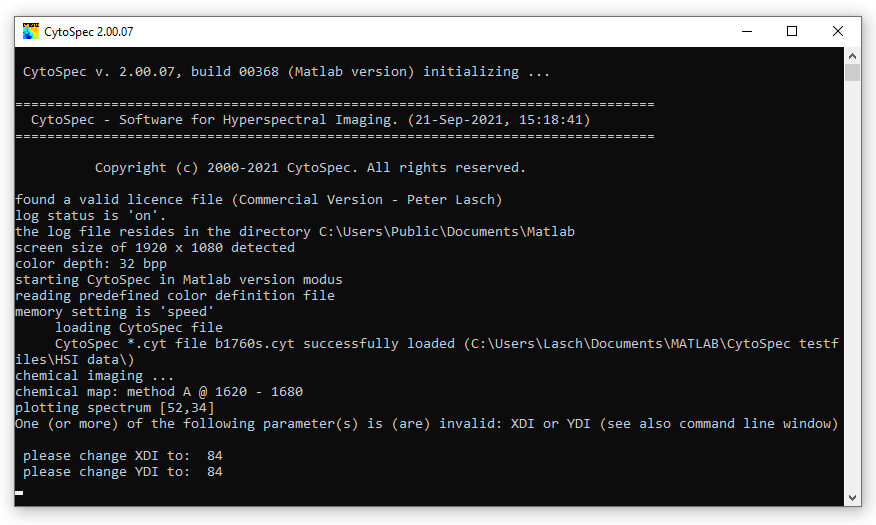
Changes of the parameters highlighted by red color are tracked and checked when leaving the 'edit HSI parameter' window. In case of errors or data inconsistencies an error messages will be displayed (see error message to the right). The CytoSpec program also makes suggestions of how to change incorrect HSI parameters (see command window below). |
|
About
This function shows, among other information, the CytoSpec program version number and the license info.
Screenshot of the dialog box 'About' with the CytoSpec software version and license information

Help - Description
This function displays the CytoSpec online help. To obtain the most recent version of the CytoSpec online help go to the CytoSpec web pages at
 https://www.cytospec.com
https://www.cytospec.com
 https://www.cytospec.com
https://www.cytospec.com
[
GENERAL |
FILE |
SPECTRAL PREPROCESSING |
SPATIAL PREPROCESSING |
UNIVARIATE IMAGING |
MULTIVARIATE IMAGING |
TOOLS |
FILE INFO |
GLOSSARY |
CONTACT: info@cytospec.com |
PUBLISHER DETAILS |
PRIVACY POLICY ]
Copyright (c) 2000-2024 CytoSpec. All rights reserved.
FILE INFO |
Copyright (c) 2000-2024 CytoSpec. All rights reserved.

 Load
Load